

Because the binding pocket of chorismate is buried deep within the protein, it may be helpful to reduce the radius of selected spheres from 10 to 6 ångstroms. Save the prepared ligand as lig_charged_tsa.mol2. The structure already comes with hydrogens, but you still need to add charges with Chimera. Thus, after removing all waters and protein atoms, select and remove chain B you are now left with the inhibitor from chain A. Also, prepare the ligand from the crystal structure that is needed for the definition of the binding pocket follow the instructions in the tutorial except that this time two inhibitor molecules carry the same name (TSA) but different chain identifiers. Follow the instructions in the tutorial, and write out the MOL2 file with hydrogens, and the PDB file without hydrogens. Prepare the receptor with UCSF Chimera starting with the 1ECM_fixed.pdb file. Save this fixed structure as a PDB file 1ECM_fixed.pdb. Click on the three hydrogen atoms to select them, then Remove Atoms in the selection labeled (sele) in the right-side toolbar. Delete the three hydrogen atoms by switching to 3 Button Viewing Mode under Mouse, and choosing Selecting Atoms mode on the right-side toolbar. Type Alt-G to build glycine residue (or select glycine from the Build menu). To prepend a residue, switch to 3 Button Editing Mode under Mouse, and click on the ring nitrogen of proline. Because it is difficult to judge what the position of the asparagine side chain in this chain should be, you will add a glycine residue. The Dock Prep tool in UCSF Chimera gets confused when a chain starts with proline, so you need to add an amino acid to the B chain before docking. In particular, chain A starts with asparagine at position 5, and chain B starts with proline at position 6. Close examination of this structure reveals that some of the residues are not visible in the crystal structure. Under File, Reinitialize PyMOL and reload structure 1ECM. In addition, you may want to analyze how docking parameters such as the grid spacing, and number of configurations generated affect the results. Follow the protocol that was outlined in the tutorial incorporating a few modifications as outlined below. You will also explore if docking supports the notion that chorismate mutase has a high affinity for the transition state of the chorismate-to-prephenate rearrangement reaction.

Analyze if docking is capable to reproduce the experimental binding geometry, and ranking of inhibitors. Your goal is to study the binding of chorismate mutase ligands to the enzyme from E. Also, rigid receptor docking to chorismate mutase would likely miss molecules that are much larger than the ligand present in the binding pocket. In situations like this, successful docking based on the rigid receptor model is possible only if the enzyme-inhibitor complex is available. Notice that the binding pocket in this protein is completely surrounded by amino acids, suggesting that the protein undergoes a significant conformational change upon ligand binding and product release. You can render the protein as a transparent surface by running the prepared script available from. Load the PDB entry 1ECM into PyMOL and examine the structure. The crystal structure of this dimeric protein ( 1ECM) is shown on the right.
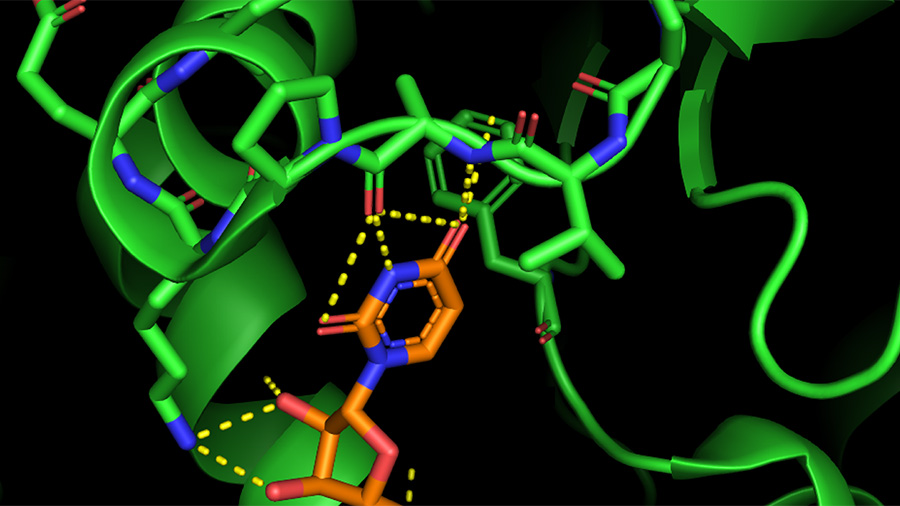
You will be testing the binding affinity of different ligands to E. Thus, it is likely that inhibitors with strong in vitro activity turn out to be weak antibacterial agents in vivo. Despite appearing as a promising antibacterial target based on its metabolic role, chorismate mutase is not well-validated experimentally. The enzyme is absent in humans mammals do not make phenylalanine via the shikimate pathway but obtain it with food. The rationale is that in the presence of such inhibitors, bacteria cannot carry out de novo synthesis of phenylalanine. It has been suggested that inhibitors of bacterial chorismate mutase may act as antibiotics (e.g. Computer-Aided Drug Design Assignments Chorismate MutaseĬhorismate mutase is involved in the biosynthesis of phenylalanine in plants and microorganisms via the shikimate pathway.


 0 kommentar(er)
0 kommentar(er)
